Computational Biology and Simulation of Biological Systems
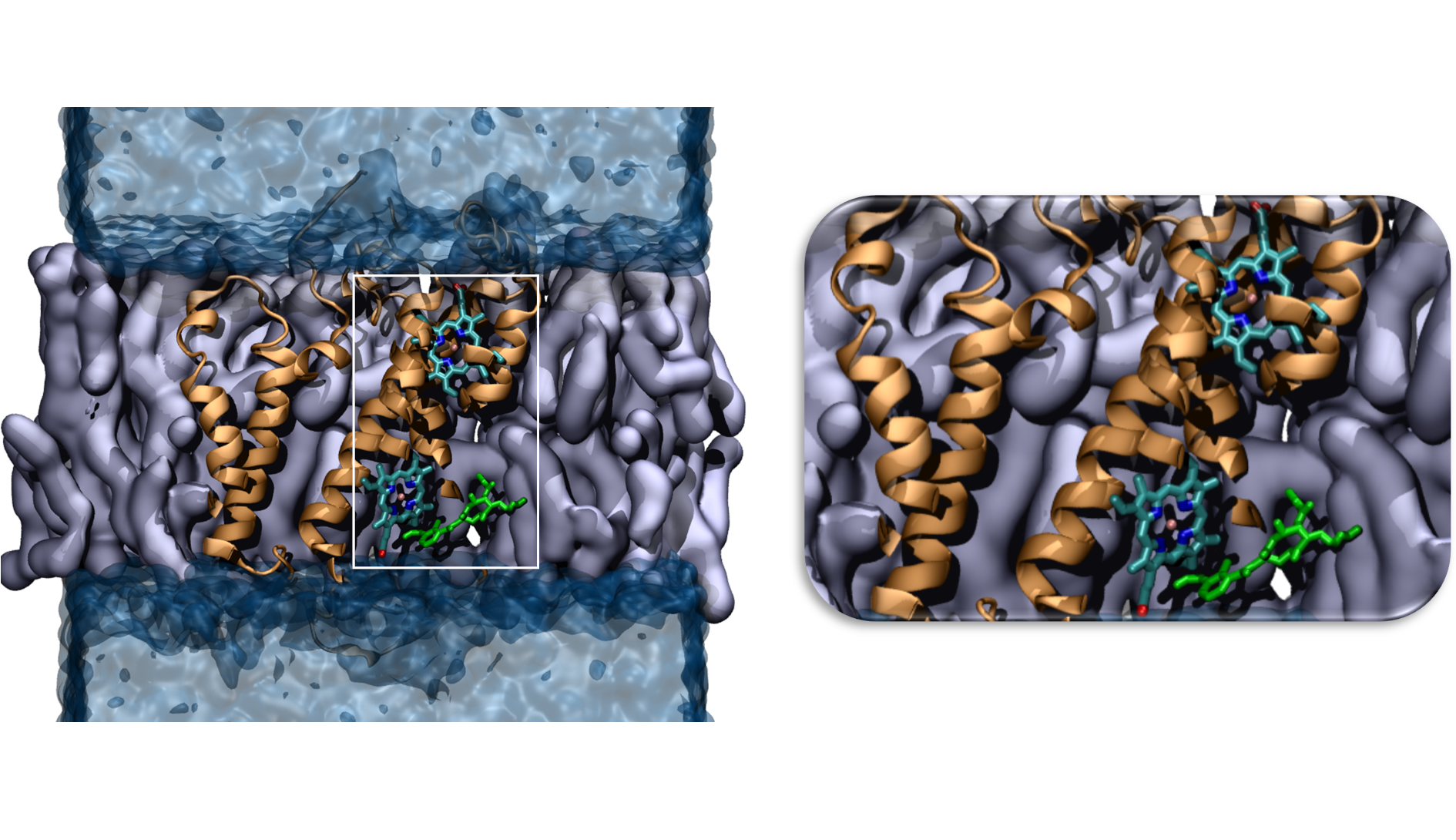
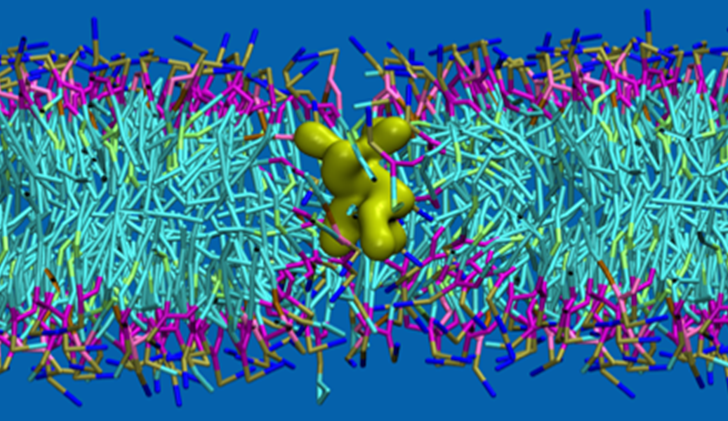
We focus on computational modeling and simulations of biological systems at molecular- and mesoscale. Structural characterization and functional mechanisms of these systems need to be elucidated, to understand diseases and develop treatments behind dysfunctions. In this framework, the main research lines are:
-
Protein folding and protein-protein interaction
-
Cell membrane penetration mechanisms of peptides
-
Computer simulations and machine learning applied in the pharmaceutical landscape
-
Signal transduction network modeling to investigate cell pathways
Key Projects
Selected Publications
-
M. Malavolta, L. Pallante, B. Mavkov, F. Stojceski, G. Grasso, A. Korfiati, S. Mavroudi, A. Kalogeras, C. Alexakos, V. Martos, D. Amoroso, G. Di Benedetto, D. Piga, K. Theofilatos & M. A. Deriu. A survey on computational taste predictors. Eur Food Res Technol. 03/2022. DOI: 10.1007/s00217-022-04044-5
-
E. Trofimenko, G. Grasso, M. Heulot, N. Chevalier, M. A Deriu, G. Dubuis, Y. Arribat, M. Serulla Llorens, S. Michel, G. Vantomme, F. Ory, J. Puyal, F. Amati, A. Lüthi, A. Danani, and C. Widmann, “Genetic, cellular and structural characterization of the membrane potential-dependent cell-penetrating peptide translocation pore”, eLife, DOI:10.7554/eLife.69832 (2021)
-
M. Serulla, G. Ichim, F. Stojceski, G. Grasso, S. Afonin, T. Schober, R. Roth, C. Godefroy, P.E. Milhiet, K. Das, A.G. Saez, A. Danani and C. Widmann, “TAT-RasGAP 317-326 kills cells by targeting inner leaflet-enriched phospholipids”, PNAS , DOI: 10.1073/pnas.2014108117 (2021)
-
G. Grasso , A. Di Gregorio , B. Mavkov, D. Piga, G. Falvo D’Urso Labate, A. Danani, and M.A. Deriu, “Fragmented blind docking: a novel protein-ligand binding prediction protocol “, Journal of Biomolecular Structure and Dynamics, 2021 Oct 12;1-10, DOI: 10.1080/07391102.2021.1988709.
-
L. Pallante, M. Malavolta, G. Grasso, A. Korfiati, S. Mavroudi, B. Mavkov, A. Kalogeras, C. Alexakos, V. Martos, D. Amoroso, G. di Benedetto, D. Piga, K. Theofilatos, M. A. Deriu. On the human taste perception: Molecular-level understanding empowered by computational methods, Trends in Food Science & Technology, 10/2021. DOI: 10.1016/j.tifs.2021.07.013
-
A. Rapallo, R. Gaspari , G. Grasso , A. Danani, “Extended Diffusion Theory: recovering dynamics from biased/accelerated molecular simulations”, Journal of Computational Chemistry, DOI: 10.1002/jcc.26474
-
S. Errico, G. Lucchesi, D. Odino, S. Muscat, C. Capitini, C. Bugelli, C. Canale, R. Ferrando, G. Grasso, D. Barbut, M. Calamai, A. Danani, M. Zasloff, A. Realini, G. Caminati, M. Vendruscolo, F. Chiti, “Making biological membrane resistant to the toxicity of misfolded protein oligomers: a lesson from trodusquemine”, Nanoscale, 2020, 12, 22596-22614, DOI: 10.1039/D0NR05285J
-
G. Grasso, A. Danani, “Molecular Simulations of Amyloid Beta Assemblies “, Advances in Physics: X, 5:1, 1770627, DOI:10.1080/23746149.2020.1770627
Contact
Prof. Andrea Danani
Professor at IDSIA